AiiDA Version 2.0 Archive Demo¶
This site is a Jupyter Notebook to demonstrate the new AiiDA v2.0 archive functionality (see aiidateam/aiida-core#5145).
The new archive gives full access to the AiiDA data exploration API,
without the need to import it into a profile, or even have a profile loaded or PostgreSQL installed.
This allows for post-computation analysis and sharing of data with colleagues,
without the usual setup overhead of AiiDA: simply install aiida-core and go!
Tip
The outputs of this notebook are dynamically generated on documentation build. Use the dropdown at the top of the page to launch interactive sessions.
from aiida import orm, __version__
__version__
/home/docs/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/aiida/manage/configuration/settings.py:51: UserWarning: Creating AiiDA configuration folder `/home/docs/.aiida`.
warnings.warn(f'Creating AiiDA configuration folder `{path}`.')
'2.0.0a1'
Reading the archive version and metadata¶
After loading the archive format class, we can read the archive version of any legacy archive.
from aiida.tools.archive import get_format
archive_format = get_format()
archive_format.latest_version
'1.0'
archive_format.read_version("archives/calc.aiida")
'1.0'
If the archive is not at the latest version, you will need to run verdi archive migrate, which will migrate the archive to the latest version.
Once the archive is at the current version, we can read the metadata of the archive.
To read data from the archive, similar to a file, we must use the open context.
import yaml
with archive_format.open("archives/calc.aiida", mode="r") as archive:
metadata = archive.get_metadata()
print(yaml.dump(metadata))
aiida_version: 2.0.0a1
compression: 6
creation_parameters:
entities_starting_set:
node:
- 05d64c2c-8f49-446f-bd07-8509d55e0c49
entity_counts:
computers: 2
links: 31
nodes: 32
users: 2
graph_traversal_rules:
call_calc_backward: true
call_calc_forward: true
call_work_backward: true
call_work_forward: true
create_backward: true
create_forward: true
input_calc_backward: true
input_calc_forward: false
input_work_backward: true
input_work_forward: false
return_backward: false
return_forward: true
include_authinfos: false
include_comments: true
include_logs: true
ctime: '2021-10-27T09:16:52.242244'
export_version: '1.0'
key_format: sha256
Querying the archive¶
We can load any entity (node, computer, user, etc.) from the archive with the get method of the read handle.
with archive_format.open("archives/calc.aiida", mode="r") as archive:
node = archive.get(orm.Node, uuid="05d64c2c-8f49-446f-bd07-8509d55e0c49")
print(node)
print(node.exit_status)
uuid: 05d64c2c-8f49-446f-bd07-8509d55e0c49 (pk: 72505) (aiida.calculations:quantumespresso.pw)
0
Note, if you try to access data from the archive outside of the context of the archive, you will get an error (unless it has already been loaded).
node.computer
---------------------------------------------------------------------------
DetachedInstanceError Traceback (most recent call last)
/tmp/ipykernel_878/3112483144.py in <module>
----> 1 node.computer
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/aiida/orm/nodes/node.py in computer(self)
324 def computer(self) -> Optional[Computer]:
325 """Return the computer of this node."""
--> 326 if self.backend_entity.computer:
327 return Computer.from_backend_entity(self.backend_entity.computer)
328
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/aiida/orm/implementation/sqlalchemy/nodes.py in computer(self)
114 """
115 try:
--> 116 return self.backend.computers.from_dbmodel(self._dbmodel.dbcomputer)
117 except TypeError:
118 return None
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/sqlalchemy/orm/attributes.py in __get__(self, instance, owner)
479 replace_context=err,
480 )
--> 481 return self.impl.get(state, dict_)
482
483
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/sqlalchemy/orm/attributes.py in get(self, state, dict_, passive)
924 return PASSIVE_NO_RESULT
925
--> 926 value = self._fire_loader_callables(state, key, passive)
927
928 if value is PASSIVE_NO_RESULT or value is NO_VALUE:
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/sqlalchemy/orm/attributes.py in _fire_loader_callables(self, state, key, passive)
960 return callable_(state, passive)
961 elif self.callable_:
--> 962 return self.callable_(state, passive)
963 else:
964 return ATTR_EMPTY
~/checkouts/readthedocs.org/user_builds/aiida-archive-demo/conda/latest/lib/python3.8/site-packages/sqlalchemy/orm/strategies.py in _load_for_state(self, state, passive, loadopt, extra_criteria)
841 return attributes.PASSIVE_NO_RESULT
842
--> 843 raise orm_exc.DetachedInstanceError(
844 "Parent instance %s is not bound to a Session; "
845 "lazy load operation of attribute '%s' cannot proceed"
DetachedInstanceError: Parent instance <DbNode at 0x7fd6adc55100> is not bound to a Session; lazy load operation of attribute 'dbcomputer' cannot proceed (Background on this error at: https://sqlalche.me/e/14/bhk3)
We can access the full QueryBuilder interface, to query data in the archive, see the querying how-to guide.
with archive_format.open("archives/calc.aiida", mode="r") as archive:
qb = archive.querybuilder()
qb.append(orm.Node, tag="calc", filters={'uuid': '05d64c2c-8f49-446f-bd07-8509d55e0c49'})
qb.append(orm.Node, with_incoming="calc")
print(qb.all())
[[<StructureData: uuid: cc4c6660-b002-456d-bb84-9bf6bf30f04e (pk: 19404)>], [<Dict: uuid: 1dea3c23-5337-4ed6-8090-637653c2fe08 (pk: 19408)>], [<TrajectoryData: uuid: bf67bdd6-e7d2-4e6e-b15d-2ae47abcfb0b (pk: 19432)>], [<Data: uuid: b17bfdf4-e9e3-4e4b-973c-e6456c9c9c9e (pk: 19411)>], [<FolderData: uuid: 1d38f6d9-74ad-4387-9570-4903009fd87b (pk: 19406)>], [<RemoteData: uuid: f5ddc748-f6c5-4b2a-ac19-149f63ea8506 (pk: 73067)>]]
Visualising the archive provenance¶
We can use the AiiDA provenance viewer to view the provenance of the archive, see the tutorial here. Note, you will need Graphviz installed for this feature.
with archive_format.open("archives/calc.aiida", mode="r") as archive:
graph = archive.graph()
graph.recurse_descendants("05d64c2c-8f49-446f-bd07-8509d55e0c49")
graph.recurse_ancestors("05d64c2c-8f49-446f-bd07-8509d55e0c49")
graph.graphviz
Analysing data from the archive¶
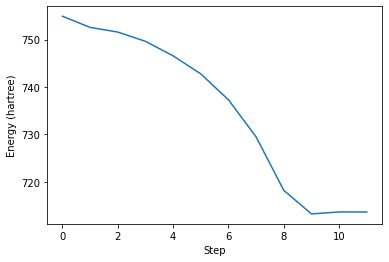
We can now plot data from the outputs of computations, for example using matplotlib.
import matplotlib.pyplot as plt
with archive_format.open("archives/calc.aiida", mode="r") as archive:
traj = archive.get(orm.Node, pk=19432)
plt.plot(traj.get_array("energy_hartree"))
ax = plt.gca()
ax.set_xlabel("Step")
ax.set_ylabel("Energy (hartree)")
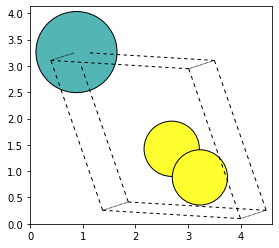
Integration with ASE is also possible, to plot atomic configurations, or even create 3D models (see ASE viewer for Jupyter notebooks).
import matplotlib.pyplot as plt
from ase.visualize.plot import plot_atoms
with archive_format.open("archives/calc.aiida", mode="r") as archive:
atoms = archive.get(orm.Node, pk=19404).get_ase()
fig, ax = plt.subplots()
ax = plot_atoms(atoms, ax, radii=0.5, rotation=("10x,10y,0z"))
from ase.visualize import view
with archive_format.open("archives/calc.aiida", mode="r") as archive:
atoms = archive.get(orm.Node, pk=19404).get_ase()
view(atoms, viewer='x3d')
Accessing repository files¶
Finally, we can access files in the repository, for example to read the raw computation log files.
with archive_format.open("archives/calc.aiida", mode="r") as archive:
folder = archive.get(orm.Node, pk=19406)
print(folder.list_object_names())
print(folder.get_object_content('aiida.out')[:988])
['_scheduler-stderr.txt', '_scheduler-stdout.txt', 'aiida.out', 'data-file.xml']
Program PWSCF v.5.2.0 starts on 25Aug2016 at 21:53:49
This program is part of the open-source Quantum ESPRESSO suite
for quantum simulation of materials; please cite
"P. Giannozzi et al., J. Phys.:Condens. Matter 21 395502 (2009);
URL http://www.quantum-espresso.org",
in publications or presentations arising from this work. More details at
http://www.quantum-espresso.org/quote
Parallel version (MPI), running on 21 processors
K-points division: npool = 21
Reading input from aiida.in
Current dimensions of program PWSCF are:
Max number of different atomic species (ntypx) = 10
Max number of k-points (npk) = 40000
Max angular momentum in pseudopotentials (lmaxx) = 3
IMPORTANT: XC functional enforced from input :
Exchange-correlation = VDW-DF2-C09 ( 1 4 16 0 2 0)
Any further DFT definition will be discarded
Please, verify this is what you really want